The RNAi-cap increases gene knockdown and decreases off-target effects
A. Introduction
Gene knockdown using siRNA or miRNA mimics is a well established method for investigation of genotypic/ phenotypic correlations in life sciences, in particular in the field of cancer research and degenerative diseases. The majority of the commercially available siRNA display overhangs located at the 3´-End of the duplex. A major challenge of this design relates to increasing potency of gene silencing and reducing off-target effects. Suppliers address these issues by improving the algorithm used to design siRNA.
Riboxx has invented a new technology termed the RNAi-cap that increases the potency of siRNA and reduces off-target effects independently from the algorithm used. In this study, the RNAi-cap technology has been applied to various siRNA from three different suppliers [A, B, and C]. The potency of gene silencing between siRNAs with or without the RNAi-cap is compared.
B. Results and discussion
B.1 The RNAi-cap technology can be applied to any siRNA or miRNA mimic
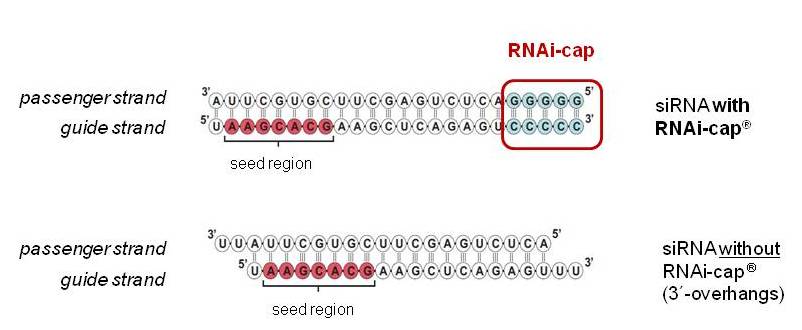
Six different siRNA's targeting different genes and different cell types were designed using different algorithms from three commercial suppliers [A, B and C], with or without the RNAi-cap. The siRNAs are displayed in the Tables below relating to each gene knockdown. As shown in Fig. 1, the RNAi-cap is located at the opposite side of the seed region of the siRNA. It can be incorporated to any siRNA designed by any algorithm, independently of the algorithm chosen. The RNAi-cap consists of a stretch of 5Cs annealed to 5Gs.
Fig. 1. Design of the siRNA with the RNAi-cap. The seed-region is shown, as well as the region bearing the RNAi-cap. The siRNA has a total length of 24 bp including the RNAi-cap.
B.2 The RNAi-cap technology strongly increases the potency of gene knockdown
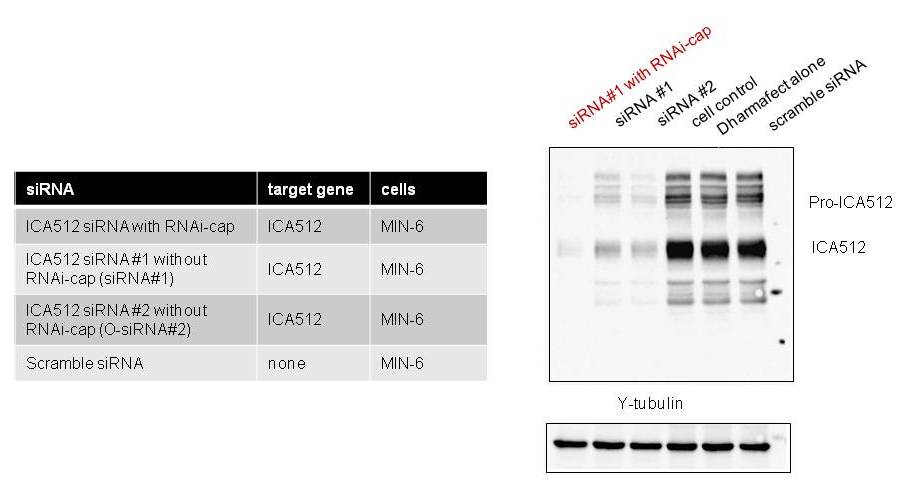
Standard siRNA (without the RNAi-cap) were purchased from commercial supplier [A], as shown in the Table below. The siRNA targeted the ICA512 gene, yielding a moderate knock down of gene expression. By adding the RNAi-cap to the siRNA, knockdown potency was increased, as evidenced in Fig. 2.
Fig. 2. The RNAi-cap strongly increases the potency of gene knockdown. Knockdown of ICA512 gene expression in MIN6 cells by siRNA bearing the RNAi-cap in comparison to two different siRNAs (siRNA#1 and siRNA #2) without the RNAi-cap designed with algorithm of supplier [A]. Expression of the ICA512 protein in MIN-6 cells was analyzed by western blot. As a control, the expression of the house-keeping gene γ-tubulin is shown.
Similarly, standard siRNA (without the RNAi-cap) were purchased from commercial supplier [B], as shown in the Table below. The siRNA targeted the caveolin-1 gene, yielding a moderate knock down of gene expression. By adding the RNAi-capto the siRNA, knock-down potency was increased, as evidenced in Fig. 3.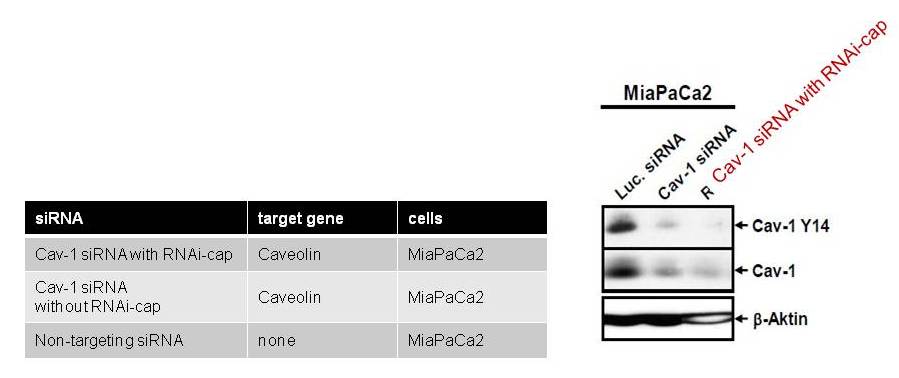 Fig. 3. The RNAi-cap strongly increases the potency of gene knockdown. Knockdown of caveolin gene expression in MIaPaCa2 cells by siRNA bearing the RNAi-cap in comparison to the same siRNA without the RNAi-cap (cav-1 siRNA) designed with algorithm of supplier [B]. Expression of the caveolin protein in MiaPaCa2 cells was performed by western blot. As a control, the expression of the house-keeping gene ß-actin is shown.
Fig. 3. The RNAi-cap strongly increases the potency of gene knockdown. Knockdown of caveolin gene expression in MIaPaCa2 cells by siRNA bearing the RNAi-cap in comparison to the same siRNA without the RNAi-cap (cav-1 siRNA) designed with algorithm of supplier [B]. Expression of the caveolin protein in MiaPaCa2 cells was performed by western blot. As a control, the expression of the house-keeping gene ß-actin is shown.
B.3 The RNAi-cap reduces off-target effects
In order to measure the reduction of off-target effects by the RNAi-cap, two siRNA targeting the TNFR-1 gene in the guide strand were used (Table below). These siRNAs are also designed to target ICAM-1 (passenger strand, hence inducing off-target effects by knocking down ICAM-1, although they should exclusively knockdown TNFR-1. The knockdown of ICAM-1 (off-target effect) by the siRNAs were compared in the presence or the absence of the RNAi-cap technology. As a control, a non-targeting siRNA (iBONi siRNA negative control form Riboxx) for both ICAM-1 or TNFR-1 was used. Expression of ICAM-1 was performed by qRT-PCR as already described (Nolte et al., 2013). As observed in Fig. 4, the siRNA without the RNAi-cap showed 50% off-target effects in comparison to a control. These were completely abolished by adding the the RNAi-cap to the duplex.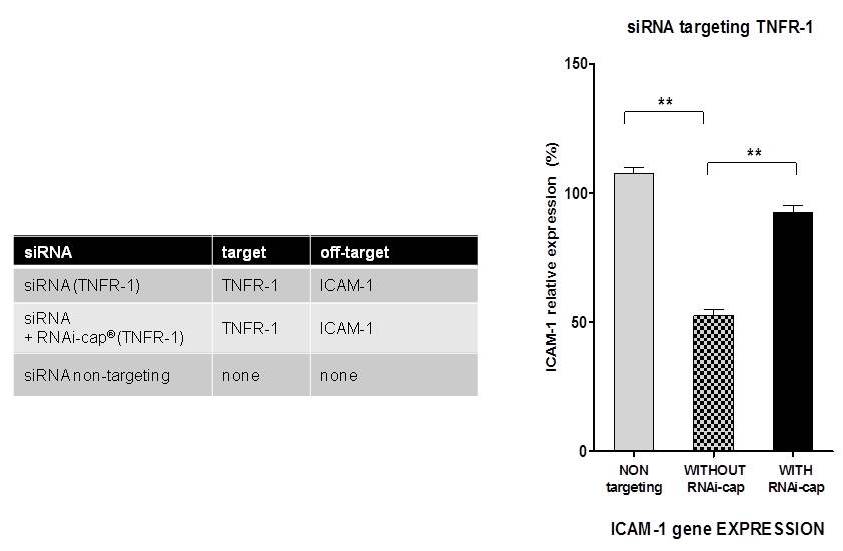
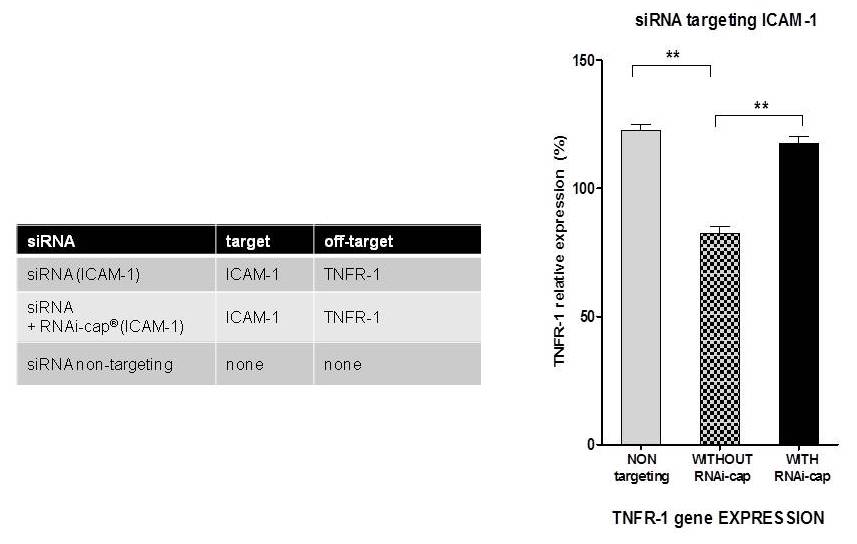
Similarly, two siRNA targeting the ICAM-1 gene in the guide strand were used (Table below). These siRNAs are also designed to target TNFR-1 (passenger strand, hence inducing off-target effects by knocking down TNFR-1, although they should exclusively knockdown ICAM-1. The knockdown of TNFR-1 (off-target effect) by the siRNAs were compared in the presence or the absence of the RNAi-cap technology. As a control, a non-targeting siRNA (iBONi siRNA negative control form Riboxx) for both ICAM-1 or TNFR-1 was used. Expression of TNFR-1 was performed by qRT-PCR as already described (Nolte et al., 2013). As observed in Fig. 5, the siRNA without the RNAi-cap® showed 40% off-target effects in comparison to a control. These were completely abolished by adding the the RNAi-cap to the duplex.
Fig. 5. Reduced off-target effects of the siRNA displaying the RNAi-cap in comparison to siRNA without the RNAi-cap designed by supplier [C]. siRNA targeting ICAM-1 without or with the RNAi-cap were compared. Expression of TNFR-1 (being the off-targer effect) was calculated by setting non-transfected, non-stimulated cells to 100% expression. As a negative control, a non-targeting siRNA (iBONi siRNA negative control) for ICAM-1 and TNFR-1 was used. Each bar represents the mean +/- standard error of mean (SEM) of n = 3. ** indicates statistical significance at a level of p < 0.01.
C. Conclusion
The RNAi-cap technology developed at Riboxx increases potency of siRNA and reduces off-target effects. The RNAi-cap technology is applicable to any siRNA designed by any algorithm, from any supplier. The RNAi-capTechnology is available at Riboxx in different siRNA and microRNA mimics formats (iVORi® siRNA, CONmiR® miRNA mimics). The RNAi-cap technology has been validated in more than 75 publications of independent scientists so far.
D. References
Nolte A, Ott K, Rohayem J, Walker T, Schlensak C, Wendel HP. Modification of small interfering RNAs to prevent off-target effects by the sense strand. N Biotechnol. 2013 Jan 25;30(2):159-65.